Frequently Asked Questions
Library Construction & Sequencing: Frequently Asked Questions
Sample Submission
What types of samples are accepted?
We accept DNA and RNA samples that are plated in our designated barcoded plates.
What are the DNA requirements for submission?
Requirements for submission vary depending on the scope of a project. Please contact us for more information.
When submitting a manifests for multiple plates, can I submit one spreadsheet, but put the different plates on separate tabs?
No, please submit one manifest spreadsheet per DNA plate.
Can I use subjects first and last names as sample IDs?
No, do not use subject names. Please do not send any information that links to subject identity.
Can I alter the submission manifest?
You may add columns, but do not remove columns or change the names of any of the columns.
There are hidden rows in the manifest, may I remove them?
No, those are hidden rows that are needed for entering the manifest into our LIMS.
May I send my samples at room temperature?
Since we do not accept samples in screw-top lids, please do not send samples at room temperature. Samples that are packed frozen and that stay frozen during delivery are much less likely to contaminate each other.
I am in the Seattle area. May I pick up my plate and bring it back when I’ve aliquoted out my DNA?
Yes, please feel free to drop by the lab. We can send instructions for how to reach us.
QC
What happens to samples that fail QC?
Samples that fail QC are not eligible to proceed into the sequencing pipeline. A single sample may be excluded from the project or in cases where the failing sample is critical to the analysis (i.e. the proband in a trio) the entire family will be excluded.
I’ve received notification that one or more of my samples has failed QC?
If a clerical error on the manifest has caused the sample to fail the sex check, you may notify the NWGC of the need to correct a manifest. If a sample has failed due to a sex conflict or due to insufficient concentration, you may submit a replacement sample by filling out a new manifest describing the replacement sample. Submission of replacement samples will delay the release of data for your project.
Sequencing
What is the turnaround time for sequencing?
Turnaround time greatly depends on the size and scope of a project and what is currently in our queue. However, on average it takes approximately 8-12 weeks after all samples in the project pass QC for data to be released. Projects with a large number of samples may require extra time for sequencing before all samples are finished.
Data Delivery
How will I know when my data are available?
You will receive an email notifying you of data release.
What data do I get back from the NWGC?
You will receive the following files: bam, bam index, multi-sample vcf, sample ID lookup table, genotype data (if applicable), and SeattleSeq annotation.
Can the NWGC provide FASTQ files in addition to bam files?
You can generate FASTQ files from the BAMs received in data release. Instructions.
Sequencing Only Projects: Frequently Asked Questions
General
What types of samples are accepted?
We accept already constructed libraries that are ready to be sequenced.
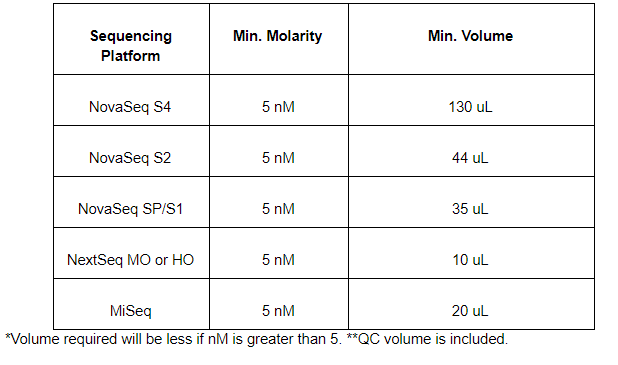
What are the volume and concentration requirements for sample submission? The minimum molarity for any library submission is 5nM, but the minimum volume depends on the flowcell type. Please see the below table:
May I send my samples at room temperature?
Please do not send samples at room temperature, unless the sample is already at room temperature and is being dropped off in person. Samples that are packed frozen and stay frozen during delivery are much less likely to contaminate each other.
I am in the Seattle area. May I drop off my sample directly to your lab?
Yes! Once the quote and sample submission form are complete and you are ready to drop off your sample, please contact us for directions and scheduling a drop off.
Can I submit custom primers with my samples?
Yes, if necessary. The amount will vary depending on the platform used, but typically 20uL of 100uM primer is sufficient.
I do not need a lot of data, can I order part of a lane?
No, we do not sell partial lanes. We recommend either choosing another platform that generates fewer reads or waiting until you have more samples to submit.
What is the turnaround time for sequencing?
Turnaround time greatly depends on the number of samples submitted and the availability of the sequencers. The typical turnaround time is 2-4 weeks for sequencing only projects.
Do you discard samples after a certain amount of time?
No. Currently, we do not have a contingency plan for discarding submitted samples for sequencing only projects.
QC
Will my samples go through quality control before they are sequenced?
Yes! All samples that are submitted as sequencing only projects require QC prior to sequencing. Each sample will be run on the Qubit and the Bioanalyzer. The QC will be reported back to the user before any sequencing begins to ensure the libraries are what is expected.
What happens if my sample fails QC?
Samples that fail QC are reported back to the user and they can reprep their libraries.
Data Delivery
How will I know when my data are available?
You will receive an email from nwgcseq@uw.edu notifying you data is available for download via Globus.
What data do I get back from the NWGC?
For Sequencing Only Projects, raw run data is converted to FASTQ format and placed on our server for you to download. If you require any raw data or run files, there is an additional bioinformatics fee based on an hourly rate.
When will I receive my data?
Our typical turnaround time is 2-4 weeks from submission to data being ready for download. This time may be shorter or longer depending on our current workload.
Globus
Do you have a Globus server endpoint?
UW has the lowest bracket of Globus service so there are limitations. Please understand that Windows Server is not an official supporter of the platform for the Globus Connect Personal (GCP) software. The Windows versions officially supported for GCP are listed in our doc here. Some users do report success using GCP on Windows Server.
Can I upload my Globus data right to AWS?
Please consider the above questions answer and troubleshoot using the link provided via Globus found here.
How long will my data be available on Globus?
Once emailed that your data is ready on Globus, you will have 120 days to access it before it is deleted indefinitely.
Genotyping Only Projects: Frequently Asked Questions
General
How long will it take to receive my results?
Sample timeline is highly dependent on project sample size and QC resolution turnaround time. Typically this ranges from 7-28 days. If there are QC fails and replacement samples are being sent, this will delay release dates. Our project managers will do our best to communicate estimated timelines with you.
QC
What happens to samples that fail QC?
Samples that fail QC are not eligible to proceed. A single sample may be excluded from the project or in cases where the failing sample is critical to the analysis (i.e. the proband in a trio) the entire family will be excluded.
What if I’ve received notification that one or more of my samples has failed QC?
If a clerical error on the manifest has caused the sample to fail the sex check, you may notify the NWGC of the need to correct a manifest.
If a sample has failed due to a sex conflict or due to insufficient concentration, you may submit a replacement sample by filling out a new manifest and describing the replacement sample. New barcoded plasticware will be sent to you for your resubmission. Submission of replacement samples will delay the start of your project.
Why did my sample fail on the genotyping/methylation assay?
Each Illumina Infinium Array kit has a typical call rate cutoff that varies based on the chip used. (generally ~98%). The Illumina data analysis software calculates and reports a detection p-value, which represents the confidence that a given transcript is expressed above the background defined by negative and positive control probes, providing a good overall QC indicator. An unusually low number of detected transcripts could result from a number of causes such as high background on the array, low signal, poor stringency or poor sample quality. This sample may not have had a similar number of transcripts detected compared to all other samples on the BeadChip, resulting in a call rate below the assay threshold.
Globus
In what format will I receive my results?
Our project managers will work with you to generate a custom final report for your samples. The file type for your results will depend on the type of genotyping assay technology selected. Raw data files (idat) may also be sent upon request.