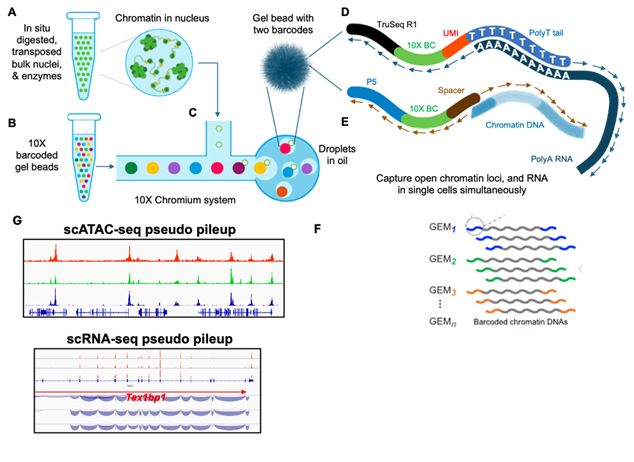
In this multiomics approach, an individual cell (or nuclei) is partitioned in one droplet (a) along with one gel bead (GEM) of millions of copies of barcoded DNA (b) and reagents as a separate picoliter reactor (c) for molecular manipulations using the 10X Chromium microfluidics system. Within a droplet, barcoded DNA is used to prime RNA and chromatin followed by linear amplification to generate cDNA- (d) and chromatin-derived fragments (e). The barcoded amplicons with droplet-specific indices can then be pooled for high-throughput sequencing (f), and the sequencing reads are processed by an established computational pipeline from which reads with identical barcodes are grouped into a distinct cell and are mapped to the reference genome to be visualized in linear browser views (g) for ATAC peaks and RNA expression.