The NWGC has built a single cell multiome data analysis pipeline (Fig. 1) that processes single cell multiome sequencing data by integrating the 10x Cell Ranger workflow, QC metrics generation, cell selection, doublet filtering to generate a variety of analyses pertaining to gene expression (GEX) and chromatin accessibility. Furthermore, since the ATAC and GEX measurements are on the very same cell, we will perform analyses that link chromatin accessibility and GEX.
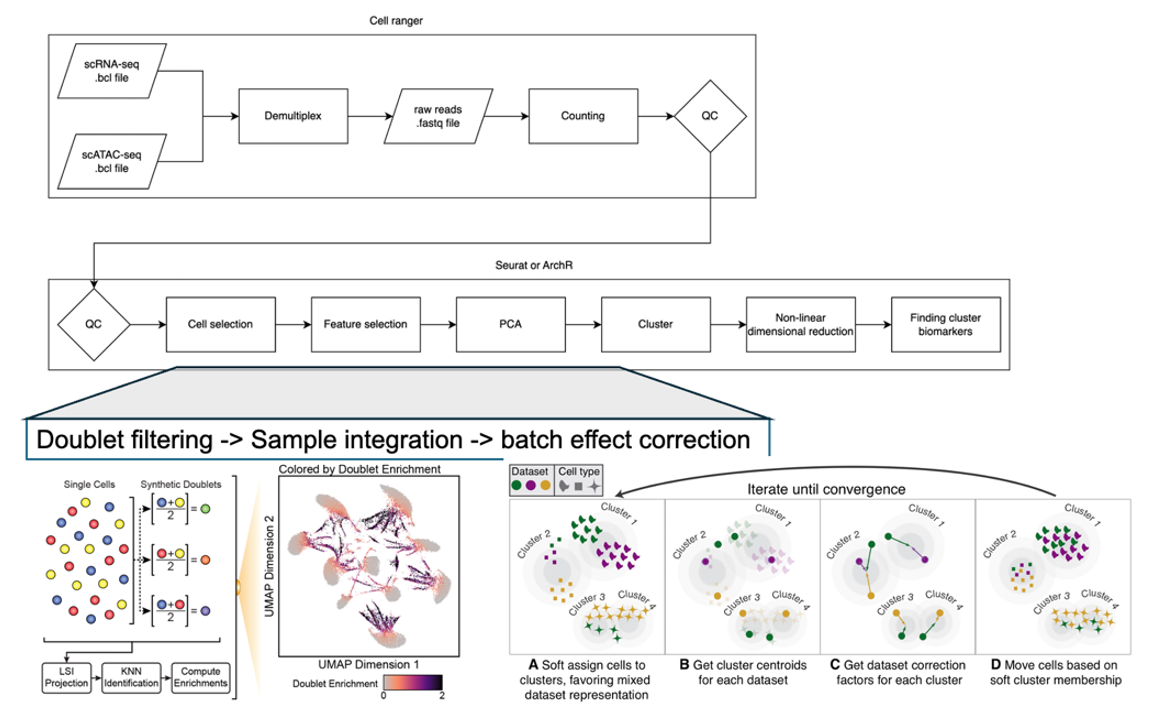
Fig.1 NWGC Multiome scRNA and scATAC analysis workflow.
We will maintain and backup data collected from our sequencing pipeline at multiple levels by storing quality-associated sequences, mapping data, and we can also provide raw reads, mapped files, QC tables, cell counts, clustering and annotation. We can release BAMs or CRAMs or FASTQs, peak files, expression level files, and metadata. We will provide all necessary computational support throughout the data dissemination process. All data will be released via our secure Globus server.